
Zhangyang Gao (高张阳)
Ph.D Candidate, Zhejiang University & Westlake University
Center for Artificial Intelligence Research and Innovation (CAIRI)
Westlake University
Location: E2-223, Westlake University, Dunyu Road #600, Hangzhou, Zhejiang, China
Email: gaozhangyang@westlake.edu.cn
[Google Scholar]
[GitHub]
[ResearchGate]
[ORCID]
Welcome to contacting me about research or internship by emails or WeChat (simpler_yet_better).
I am eagerly seeking a postdoctoral position beginning in August 2025!
News
- [2025.2] One paper on, De Novo Mass Spectrometry Peptide Sequencing, has been accepted by ICLR 2025 , congrats to Shaorong Chen.
- [2025.2] One paper on, Protein Motif-Scaffolding, has been accepted by ICLR 2025 , congrats to Yufei Huang.
- [2025.2] One paper on, Protein Post-Translational Modification Prediction, has been accepted by ICLR 2025 , congrats to Cheng Tan.
- [2024.10] One paper on, Protein language, has been accepted by AAAI 2025 , congrats to Cheng Tan.
- [2024.10] One paper on, Antibody design, has been accepted by AAAI 2025 (Oral) , congrats to Cheng Tan.
- [2024.10] One paper on, Antibody design, has been accepted by AAAI 2025 , congrats to Lirong Wu.
- [2024.09] One paper on, Protein sequence design, has been accepted by NeurIPS 2024
- [2024.09] One paper on, Protein representation learning, has been accepted by NeurIPS 2024, congrats to Bozhen Hu.
- [2024.09] One paper on, Peptide Sequencing in Proteomics, has been accepted by NeurIPS 2024, congrats to Jun Xia.
- [2024.05] One paper on, Self-supervised Graph Language Modeling, has been accepted by ICML 2024
- [2024.05] One paper on, Molecule Docking, has been accepted by ICML 2024, congrats to Yufei Huang.
- [2024.03] One paper on, Protein Sequence Design, has been accepted by ICLR 2024
- [2024.03] One paper on, graph distillation, has been accepted by TKDE, congrats to Lirong Wu.
- [2024.03] One paper on, Medical Image Representation Learning, has been accepted by CVPR 2024, congrats to Zhe Li.
- [2024.03] One paper on, point cloud pretraining, has been accepted by CVPR 2024, congrats to Zhe Li.
- [2024.03] One paper on, self-supervised graph learning, has been accepted by ICDE 2024, congrats to Jun Xia.
- [2024.02] One survey on, diffusion model, has been accepted by TKDE, congrats to Hanqun Cao.
- [2023.12] One paper on, compound-protein interaction, has been accepted by TKDE, congrats to Lirong Wu.
- [2023.12] One paper on, Molecule Representation Learning, has been accepted by AAAI 2024, congrats to Yufei Huang.
- [2023.12] One paper on, antibody design, has been accepted by AAAI 2024, congrats to Cheng Tan.
- [2023.09] One paper on, Protein Sequence Design, has been accepted by NeurIPS 2023
- [2023.09] One paper on, video prediction, has been accepted by NeurIPS 2023, congrats to Cheng Tan.
- [2023.06] One paper on, molecule pretraining, has been accepted by ECML 2023.
- [2023.06] One paper on, molecule generation, has been accepted by ECML 2023, congrats to Cheng Tan.
- [2023.03] One paper on, Protein Sequence Design, has been accepted by ICLR 2023 (Spotlight)
- [2023.03] One paper on, molecule pretraining, has been accepted by ICLR 2023, congrats to Jun Xia.
- [2023.03] One paper on, protein sequence design, has been accepted by ICASSP 2023, congrats to Cheng Tan.
- [2023.03] One paper on, video prediction, has been accepted by CVPR 2023, congrats to Cheng Tan.
- [2023.01] One paper on, graph learning, has been accepted by TNNLS, congrats to Lirong Wu.
- [2022.03] One paper on, Video Prediction, has been accepted by CVPR 2022
- [2022.03] One paper on, semi-supervised learning, has been accepted by CVPR 2022, congrats to Cheng Tan.
- [2021.12] One survey on, graph self-supervised learning, has been accepted by TKDE, congrats to Lirong Wu.
- [2021.12] One paper on, spatio-temporal prediction, has been accepted by AAAI 2021, congrats to Haitao Lin.
Research Interest
Currently, I focus on the following research topics:- Video Prediction
- AI4Science (Molecule Modeling, Protein Design)
Education
- 2020.09-present Ph.D in CAIRI, Westlake University. Supervisor: Prof. Stan Z. Li
- 2016.09-2020.06 B.E. in School of Automation, Central South University. Ranks first in the major and college.
Publications
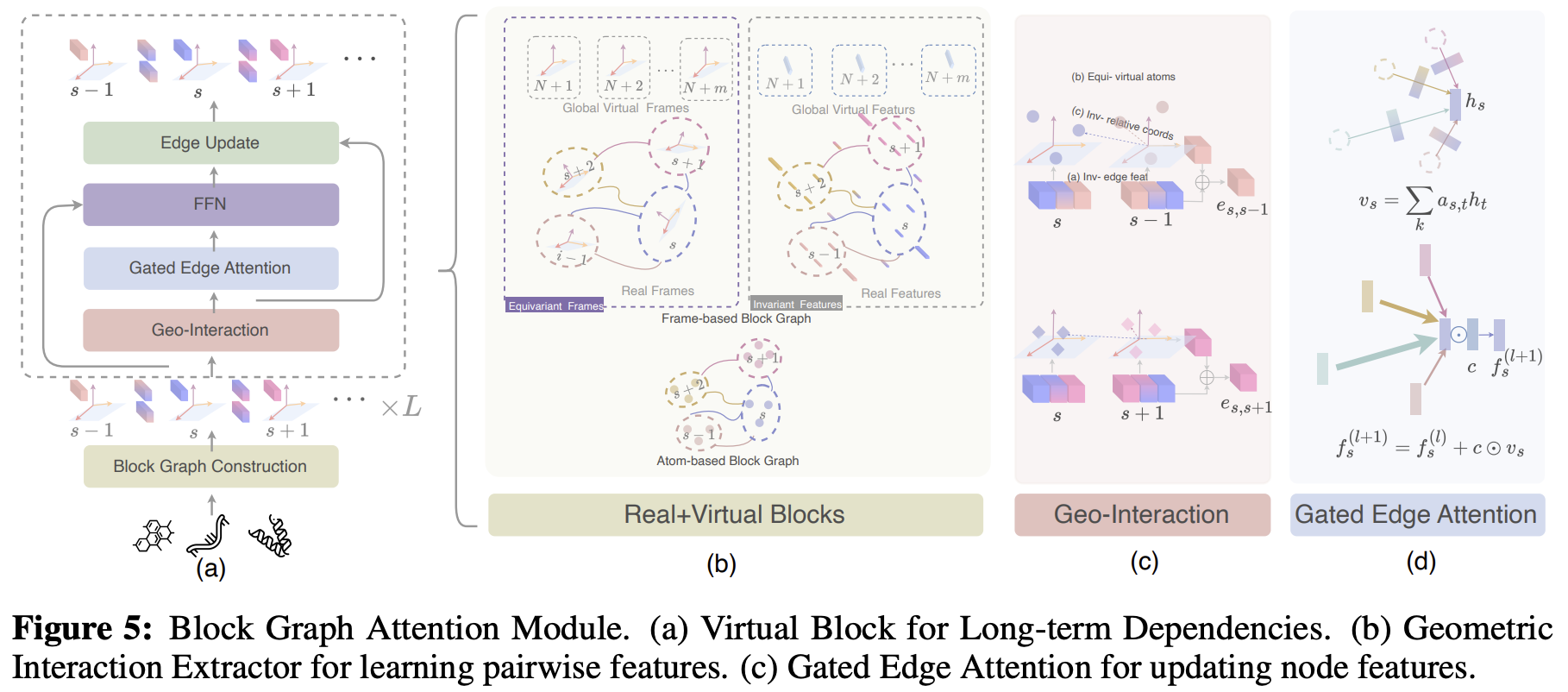
UniIF: Unified Molecule Inverse Folding
Zhangyang Gao, Jue Wang, Cheng Tan, Lirong Wu, Yufei Huang, Siyuan Li, Zhirui Ye, Stan Z. Li
NeurIPS, 2024
[PDF]
[Github]
[BibTeX]
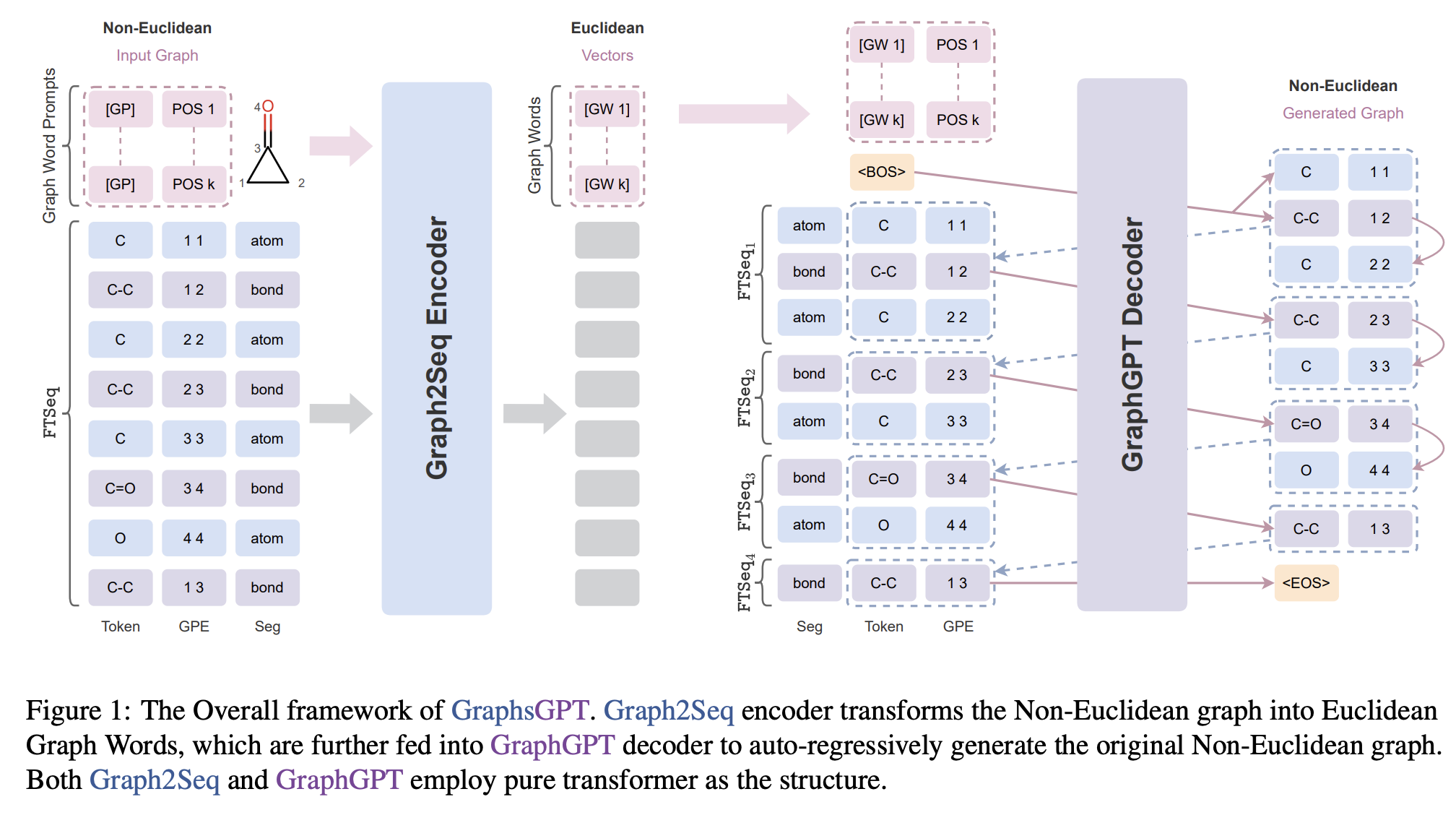
A Graph is Worth $ K $ Words: Euclideanizing Graph using Pure Transformer
Zhangyang Gao, Daize Dong, Cheng Tan, Jun Xia, Bozhen Hu, Stan Z.Li
ICML, 2024
[PDF]
[Github]
[BibTeX]
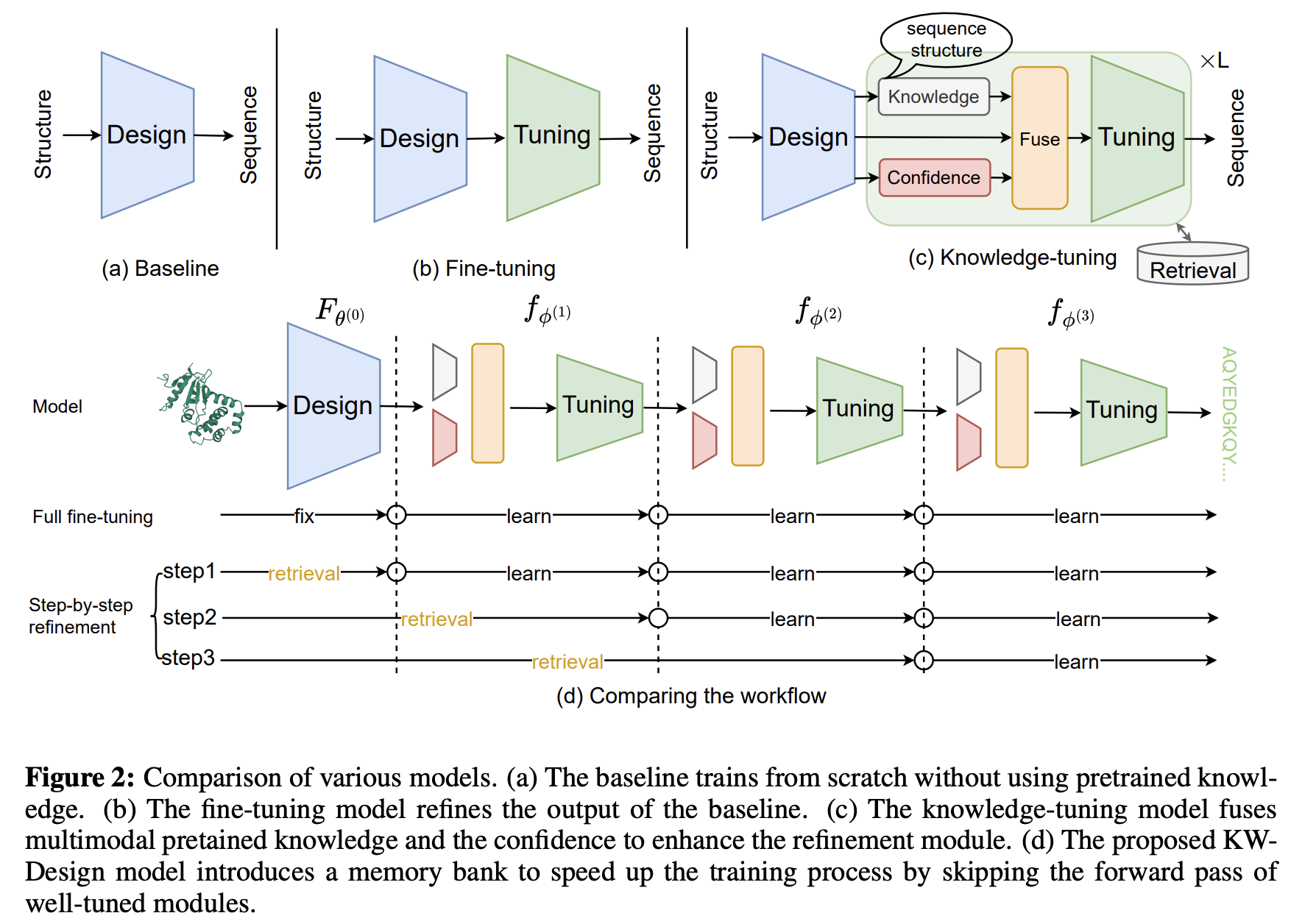
KW-Design: Pushing the Limit of Protein Design via Knowledge Refinement
Zhangyang Gao, Cheng Tan, Xingran Chen, Yijie Zhang, Jun Xia, Siyuan Li, Stan Z.Li
ICLR, 2024
[PDF]
[Github]
[BibTeX]
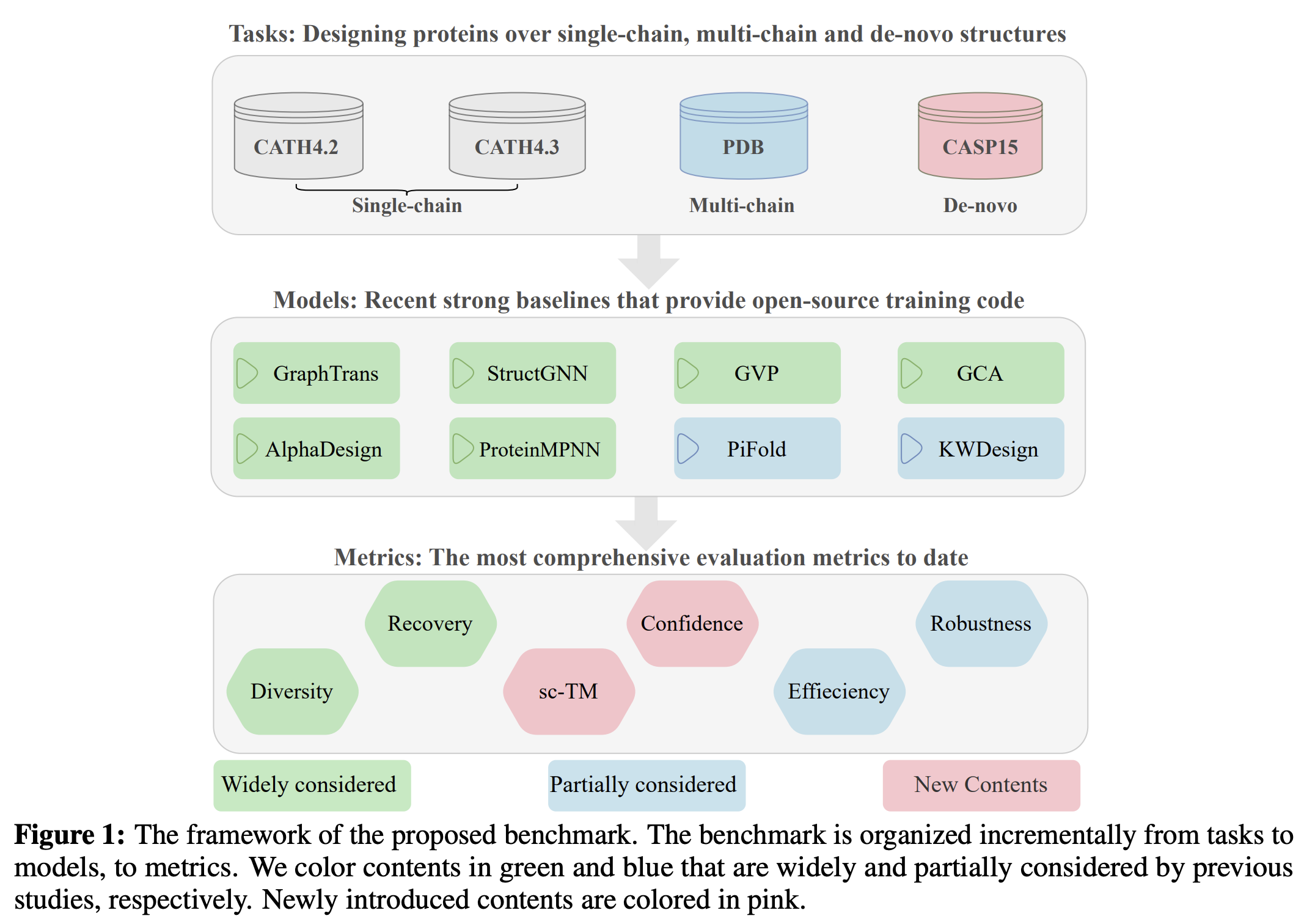
Proteininvbench: Benchmarking protein inverse folding on diverse tasks, models, and metrics
Zhangyang Gao, Cheng Tan, Yijie Zhang, Xingran Chen, Lirong Wu, Stan Z.Li
NeurIPS, 2023
[PDF]
[Github]
[BibTeX]
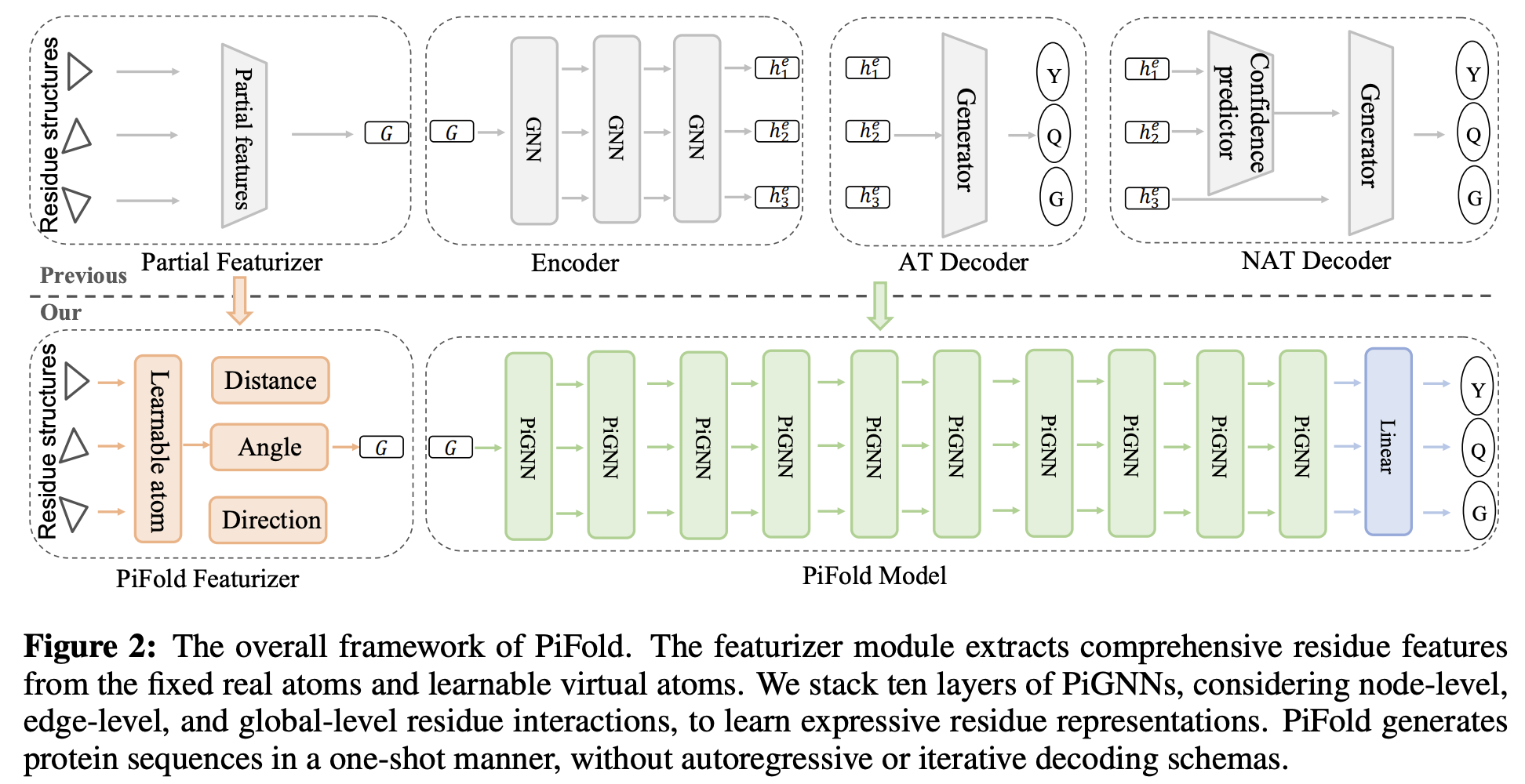
PiFold: Toward effective and efficient protein inverse folding
Zhangyang Gao, Cheng Tan, Stan Z.Li
ICLR, 2023 (Spotlight)
[PDF]
[Github]
[BibTeX]
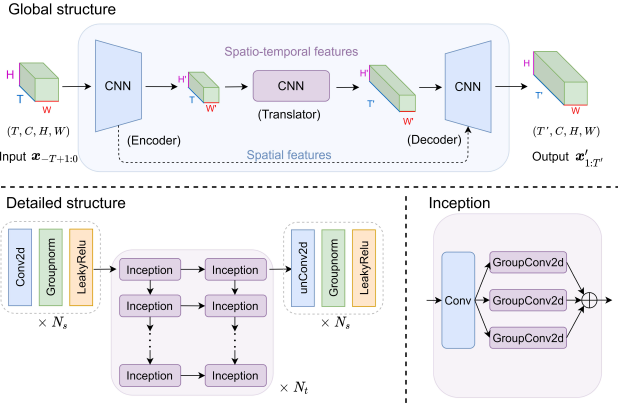
Simvp: Simpler yet better video prediction
Zhangyang Gao, Cheng Tan, Lirong Wu, Stan Z.Li
CVPR, 2022
[PDF]
[Github]
[BibTeX]
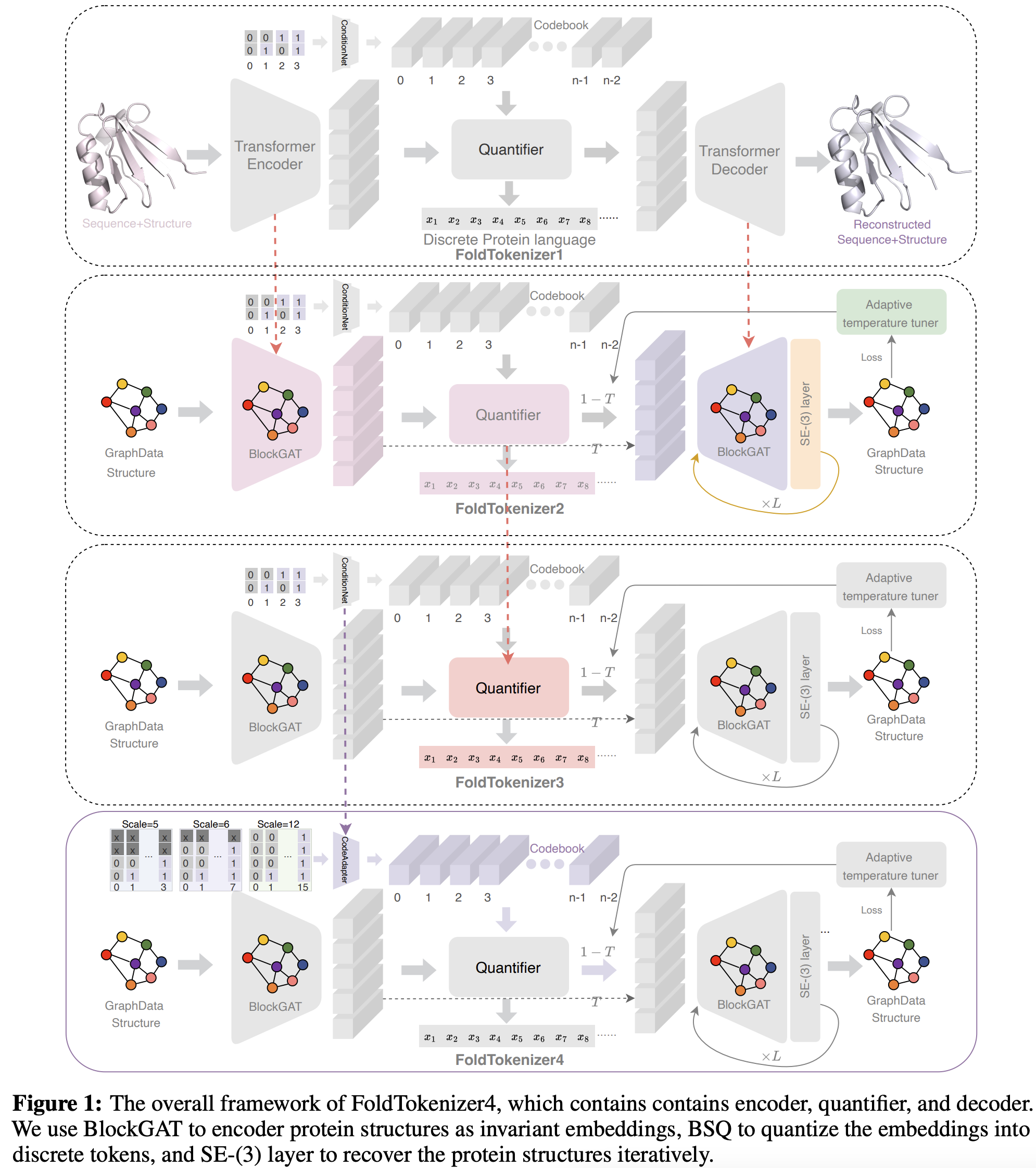
Foldtoken: Learning protein language via vector quantization and beyond
Zhangyang Gao, Cheng Tan, Jue Wang, Yufei Huang, Lirong Wu, Stan Z.Li
AAAI, 2025
[PDF]
[BibTeX]
FoldToken2: Learning compact, invariant and generative protein structure language
Zhangyang Gao, Cheng Tan, Stan Z.Li
Arxiv, 2024
[PDF]
[Github]
[BibTeX]
FoldToken3: Fold Structures Worth 256 Words or Less
Zhangyang Gao, Cheng Tan, Stan Z.Li
Arxiv, 2024
[PDF]
[BibTeX]
FoldToken4: Consistent & Hierarchical Fold Language
Zhangyang Gao, Cheng Tan, Stan Z.Li
Arxiv, 2024
[PDF]
[BibTeX]
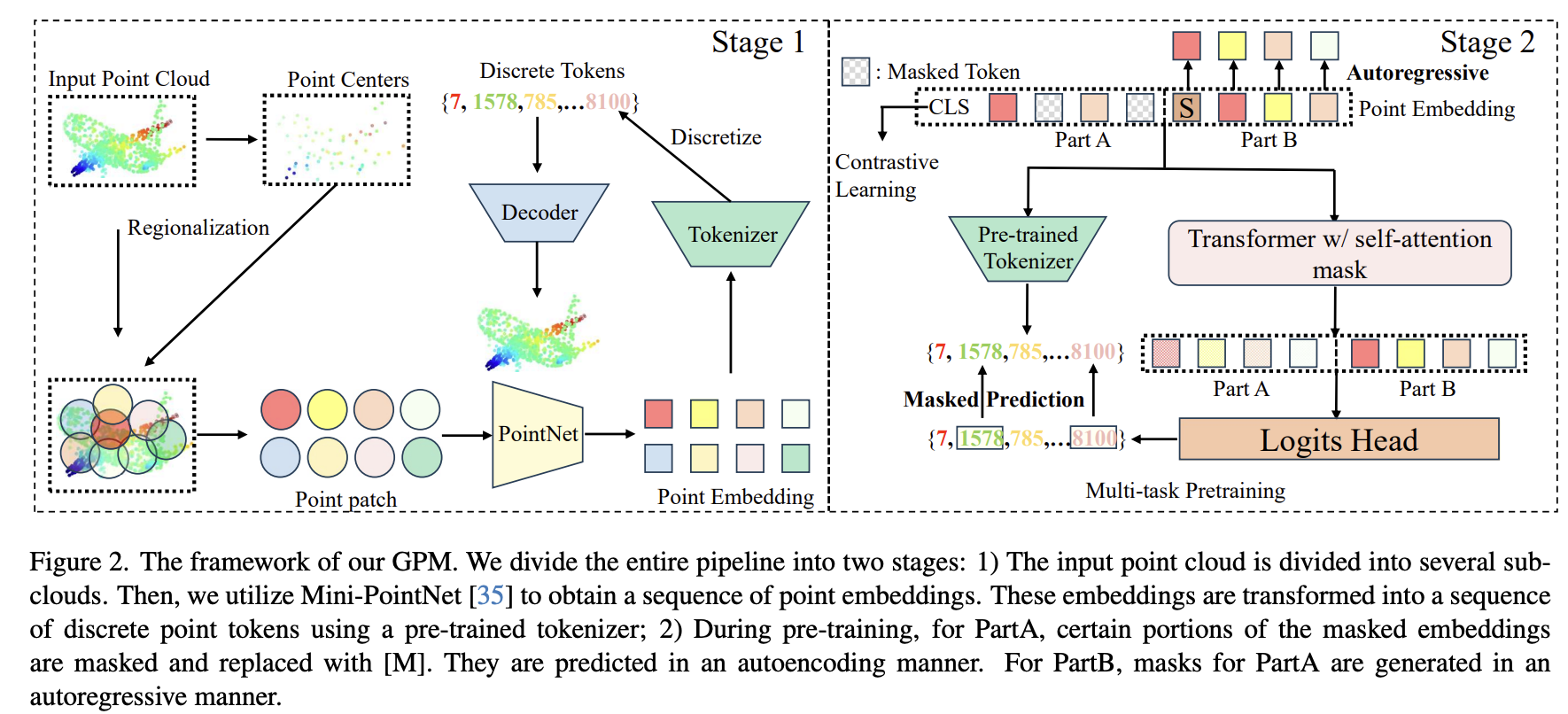
General Point Model Pretraining with Autoencoding and Autoregressive
Zhe Li, Zhangyang Gao*, Cheng Tan, Bocheng Ren, Laurence T Yang, Stan Z.
CVPR, 2024
[PDF]
[BibTeX]
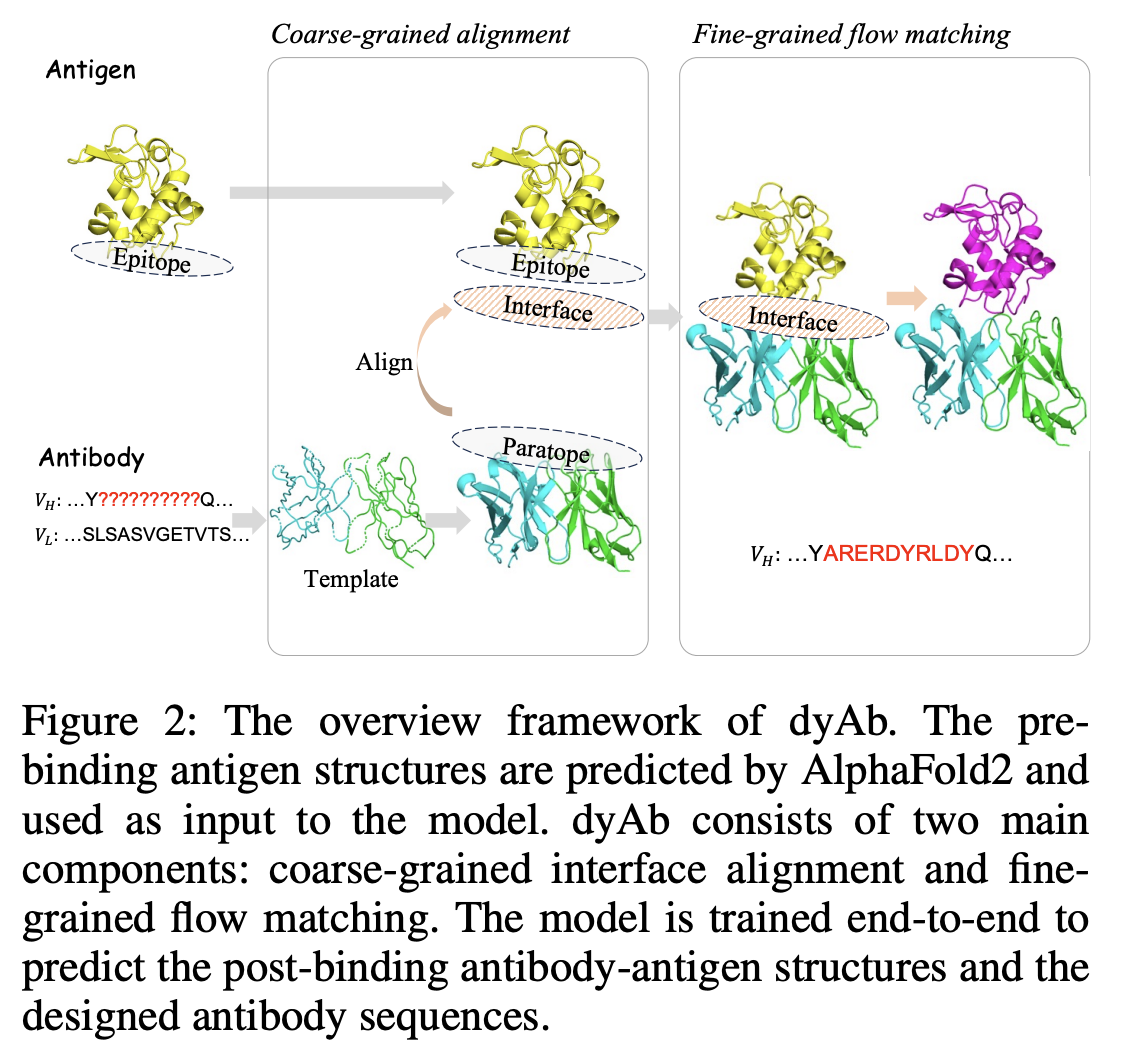
dyAb: Flow Matching for Flexible Antibody Design with AlphaFold-driven Pre-binding Antigen
Cheng Tan, Yijie Zhang, Zhangyang Gao, Yufei Huang, Haitao Lin, Lirong Wu, Fandi Wu, Mathieu Blanchette, Stan Z. L
AAAI, 2025 (Oral)
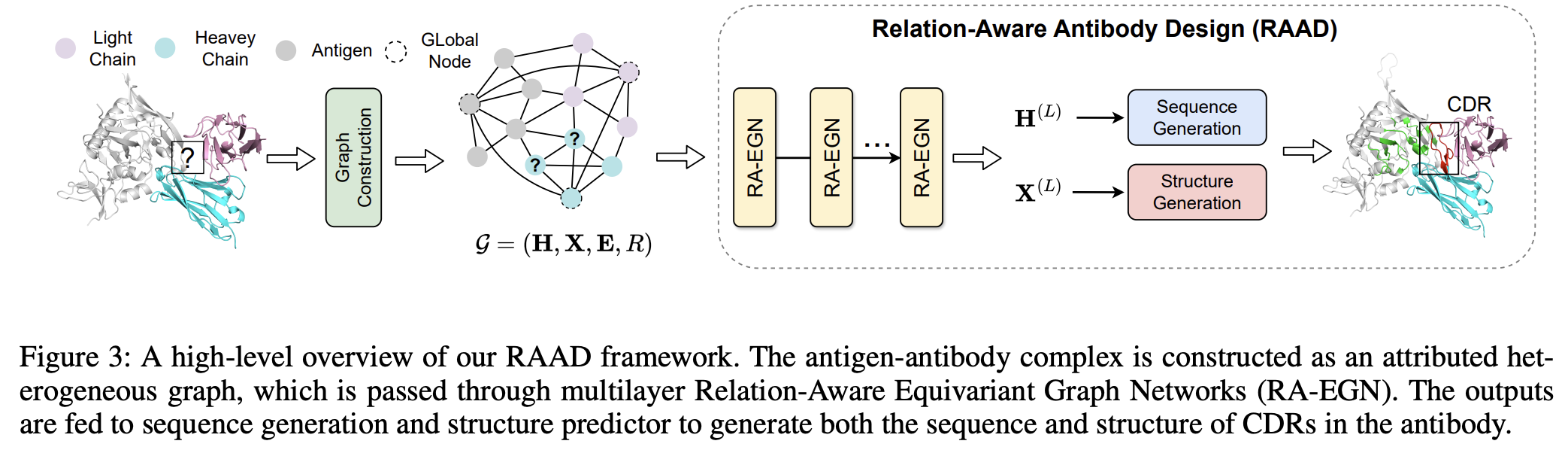
Relation-Aware Equivariant Graph Networks for Epitope-Unknown Antibody Design and Specificity Optimization
Lirong Wu, and Haitao Lin, Yufei Huang and Zhangyang Gao and Cheng Tan, Yunfan Liu, Tailin Wu, and Stan Z. Li
AAAI, 2025
[BibTeX]
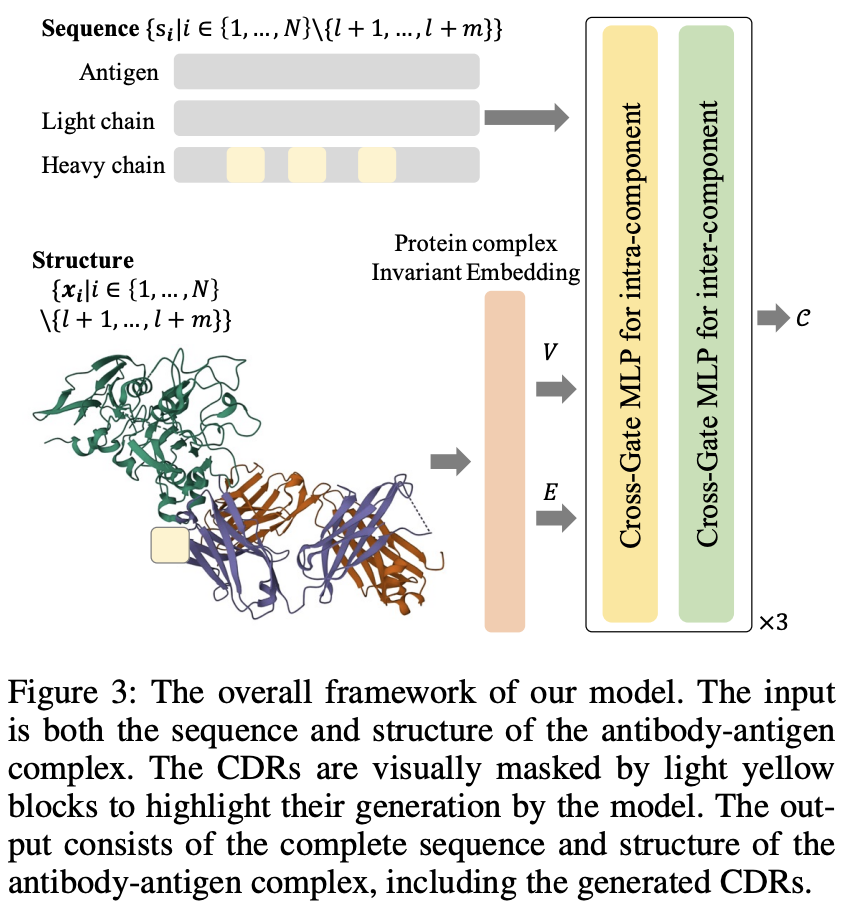
Cross-Gate MLP with Protein Complex Invariant Embedding Is a One-Shot Antibody Designer
Cheng Tan, Zhangyang Gao*, Lirong Wu, Jun Xia, Jiangbin Zheng, Xihong Yang, Yue Liu, Bozhen Hu, Stan Z.
AAAI, 2024
[PDF]
[Github]
[BibTeX]
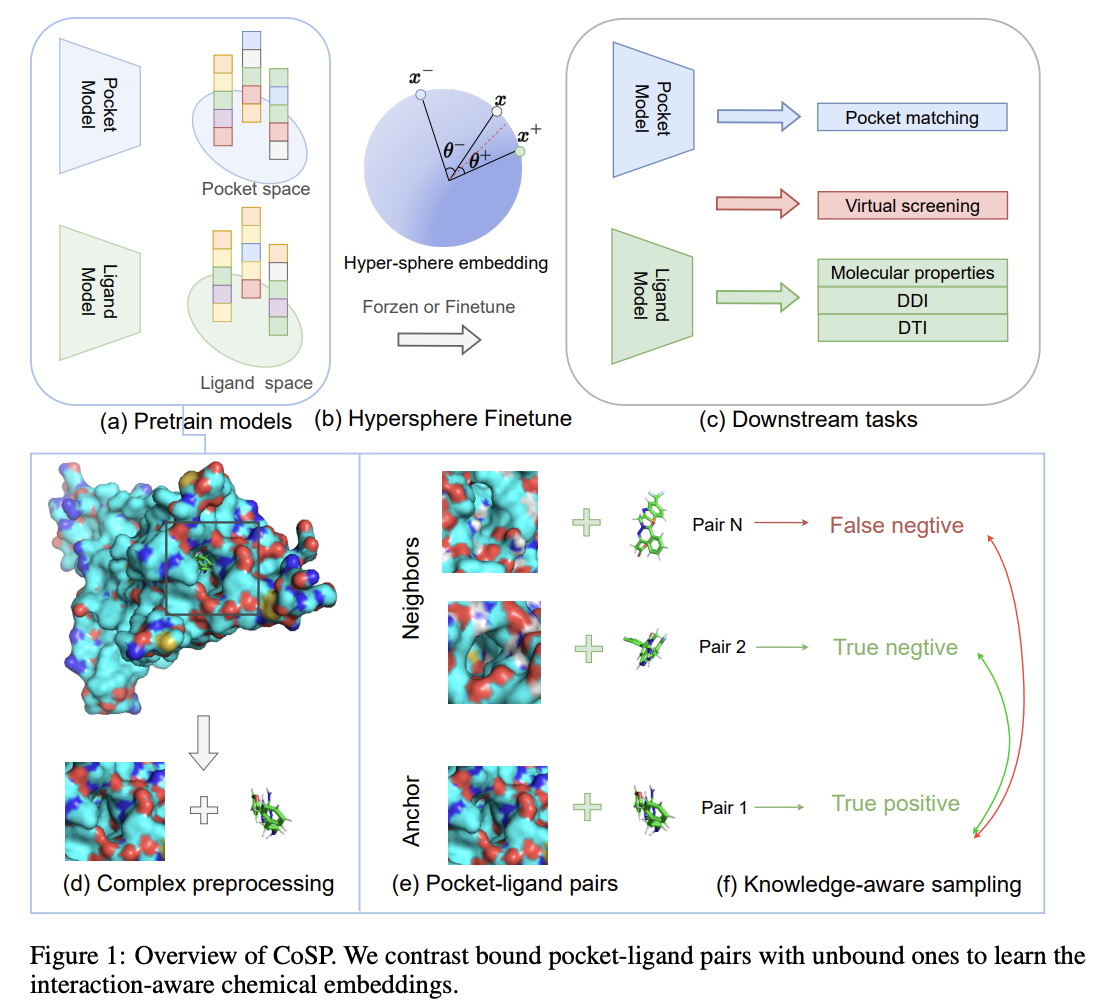
Co-supervised Pre-training of Pocket and Ligand
Zhangyang Gao, Cheng Tan, Jun Xia, Stan Z.
ECML, 2023
[PDF]
[BibTeX]
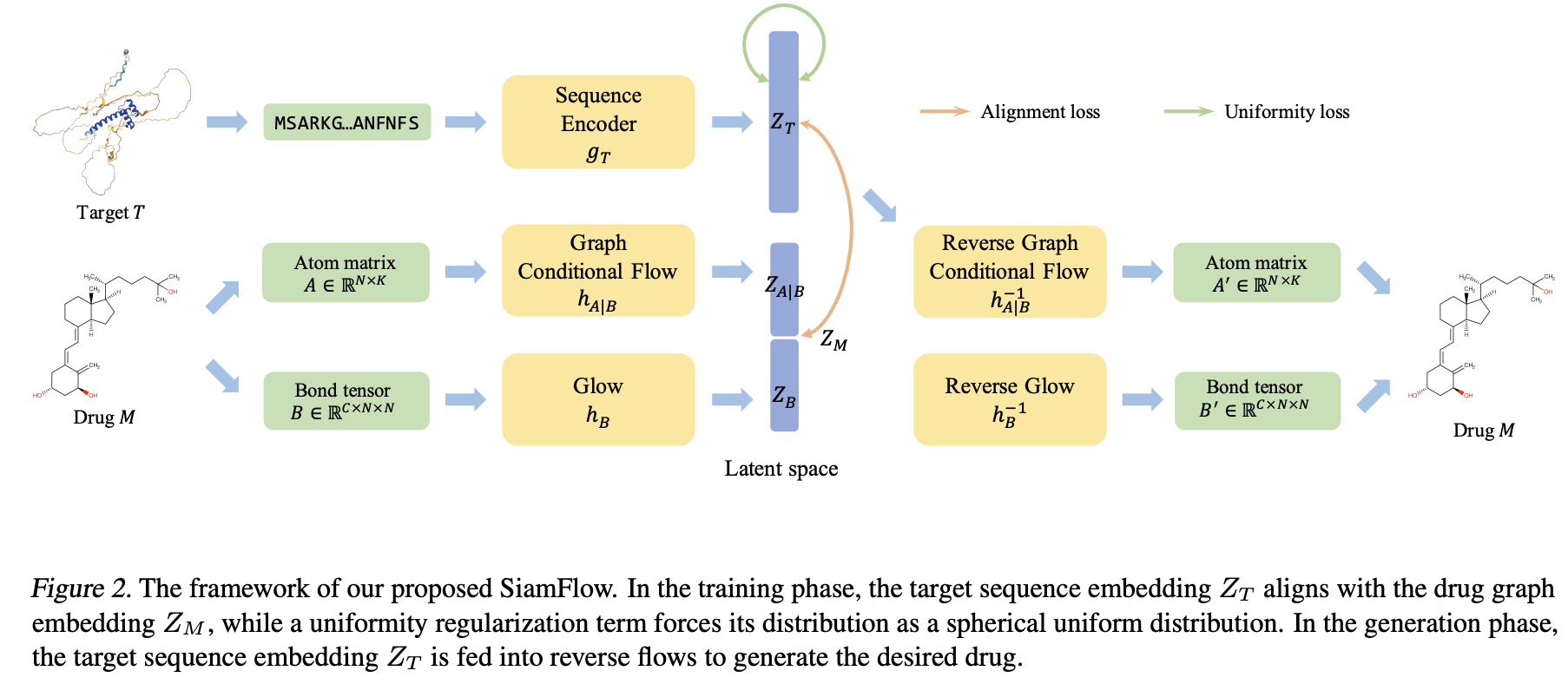
Target-aware molecular graph generation
Cheng Tan, Zhangyang Gao*, Stan Z.
ECML, 2023
[PDF]
[BibTeX]
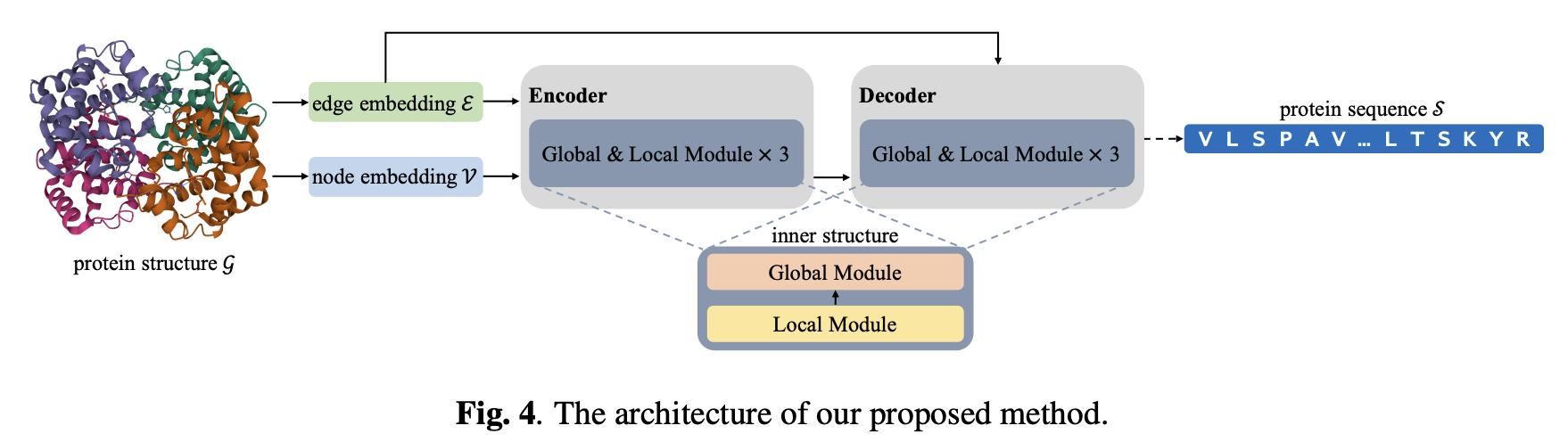
Global-context aware generative protein design
Cheng Tan, Zhangyang Gao*, Jun Xia, Bozhen Hu, Stan Z.
ICASSP, 2023
[PDF]
[Github]
[BibTeX]
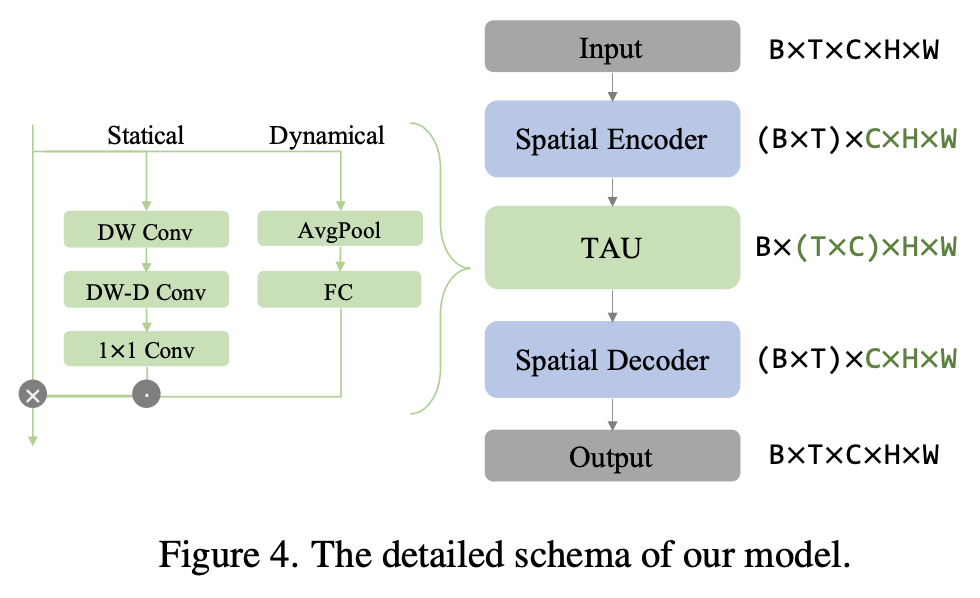
Temporal attention unit: Towards efficient spatiotemporal predictive learning
Cheng Tan, Zhangyang Gao*, Lirong Wu, Yongjie Xu, Jun Xia, Siyuan Li and Li, Stan Z.
CVPR, 2023
[PDF]
[Github]
[BibTeX]
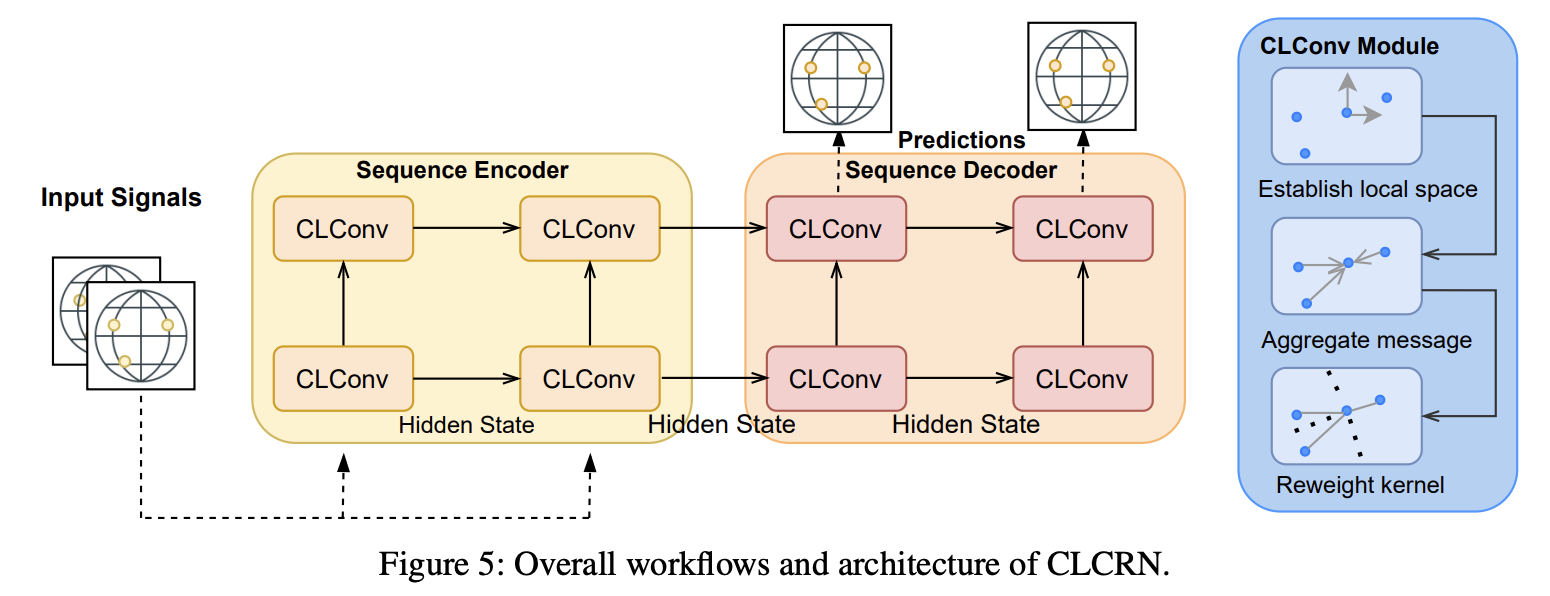
Conditional local convolution for spatio-temporal meteorological forecasting
Haitao Lin, Zhangyang Gao*, Yongjie Xu, Ling Li, Stan Z.Li
AAAI, 2021
[PDF]
[Github]
[BibTeX]
Hyperspherical consistency regularization
Cheng Tan, Zhangyang Gao*, Lirong Wu, Siyuan Li, Stan Z.Li
CVPR, 2022
[PDF]
[Github]
[BibTeX]
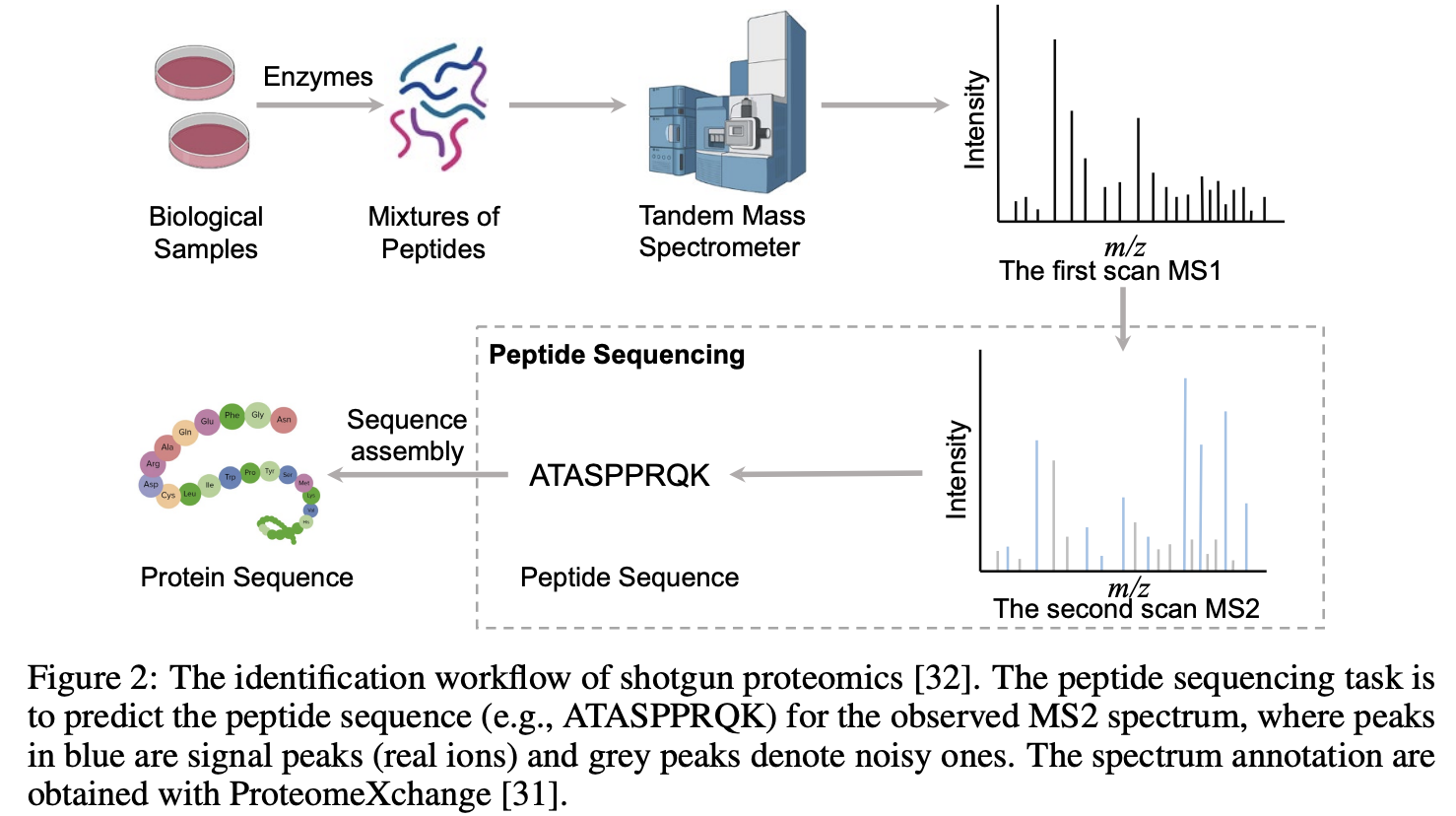
Towards Robust De Novo Peptide Sequencing in Proteomics against Data Biases
Jun Xia, Shaorong Chen, Jingbo Zhou, Xiaojun Shan, Wenjie Du, Zhangyang Gao, Cheng Tan, Bozhen Hu, Jiangbin Zheng, Stan Z. Li
NeurIPS, 2024
[PDF]
[BibTeX]
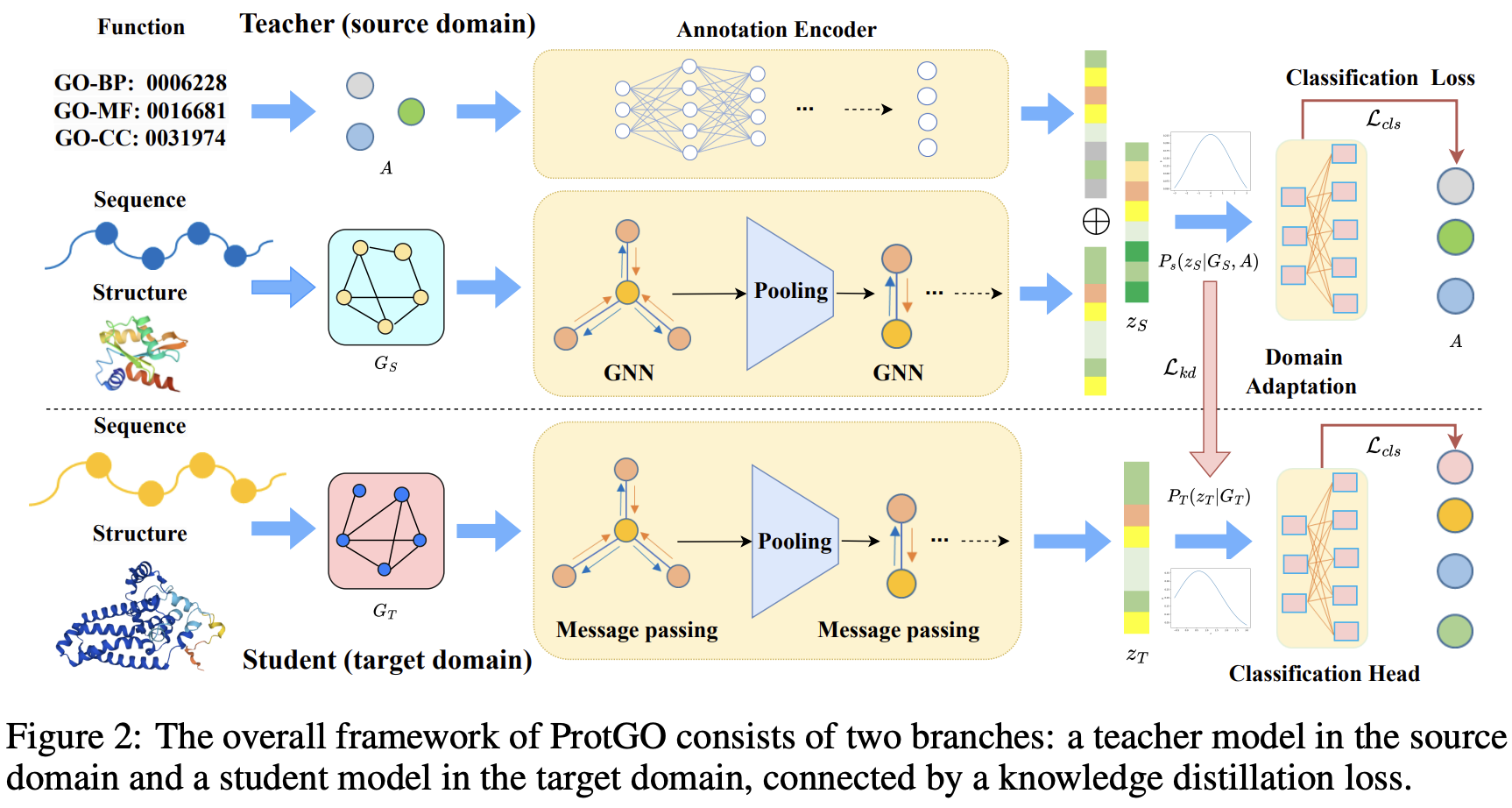
ProtGO: Function-Guided Protein Modeling for Unified Representation Learning
Bozhen Hu, Cheng Tan, Yongjie Xu, Zhangyang Gao, Jun Xia, Lirong Wu, Stan Z. Li
NeurIPS, 2024
[PDF]
[BibTeX]
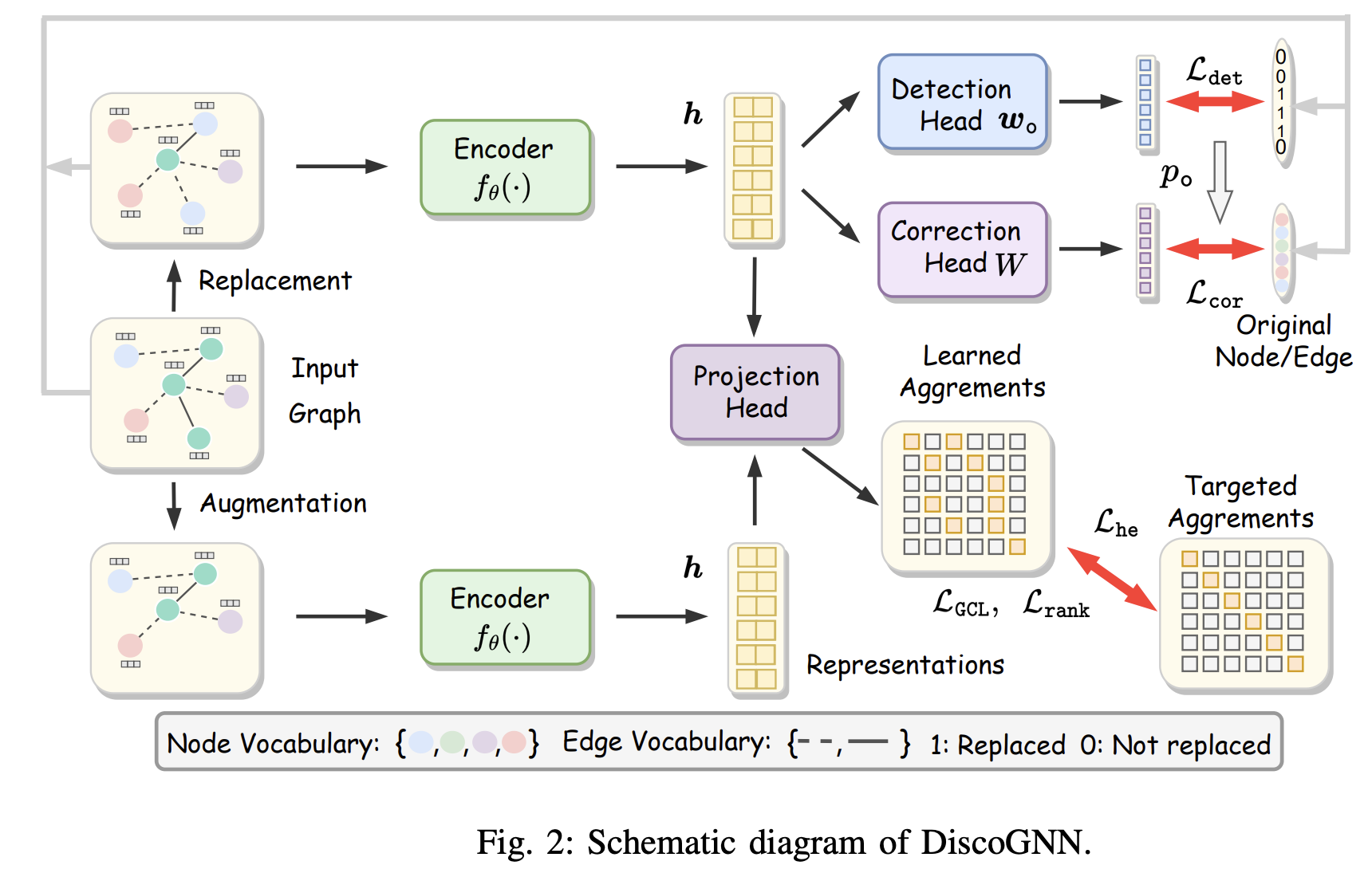
DiscoGNN: A Sample-Efficient Framework for Self-Supervised Graph Representation Learning
Jun Xia, Shaorong Chen, Yue Liu, Zhangyang Gao, Jiangbin Zheng, Xihong Yang, Stan.Z
ICDE, 2024
[PDF]
[BibTeX]
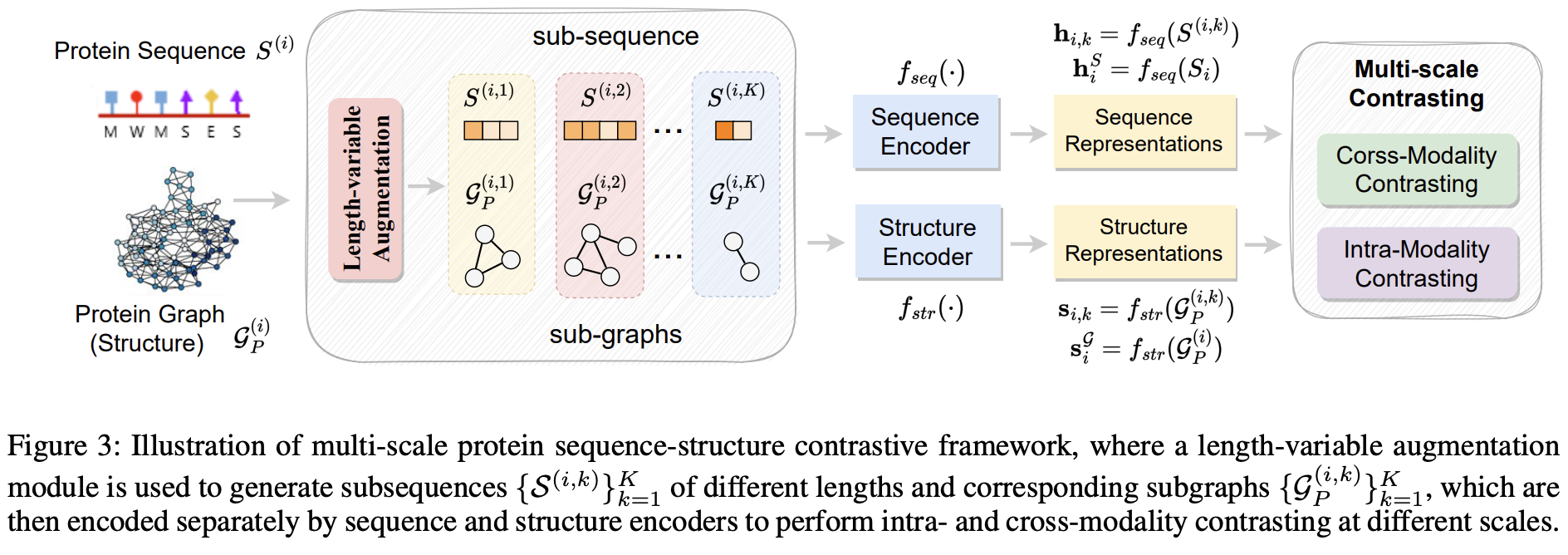
PSC-CPI: Multi-Scale Protein Sequence-Structure Contrasting for Efficient and Generalizable Compound-Protein Interaction Prediction
Lirong Wu, Yufei Huang, Chen Tan, Zhangyang Gao, Bozhen Hu, Haitao Lin, Zicheng Liu, Stan.Z
AAAI, 2024
[PDF]
[Github]
[BibTeX]
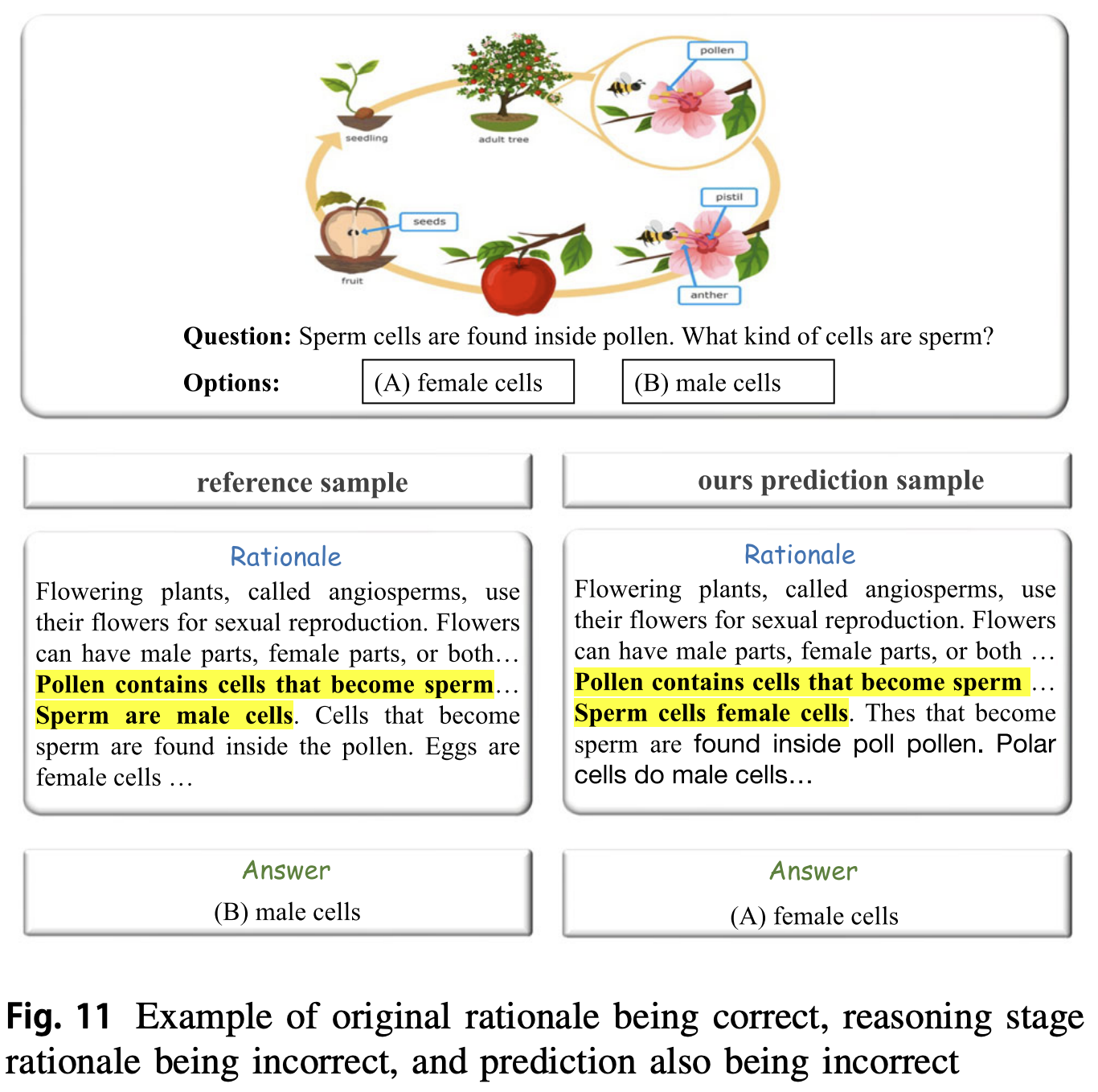
Enhancing human-like multimodal reasoning: a new challenging dataset and comprehensive framework
Jingxuan Wei, Cheng Tan, Zhangyang Gao, Linzhuang Sun, Siyuan Li, Bihui Yu, Ruifeng Guo & Stan.Z
Neural Computing and Applications, 2024
[PDF]
[BibTeX]
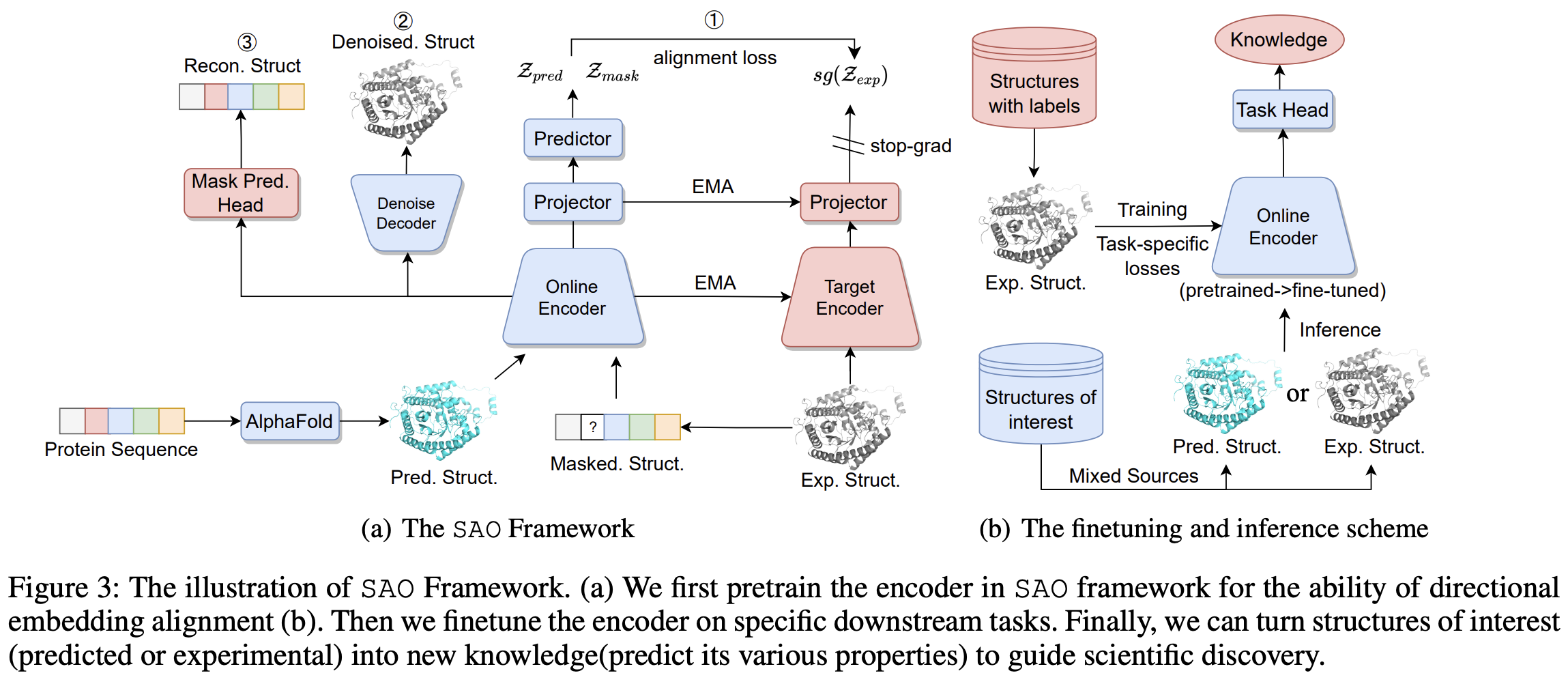
Protein 3D Graph Structure Learning for Robust Structure-Based Protein Property Prediction
Yufei Huang, Siyuan Li, Lirong Wu, Jin Su, Haitao Lin, Odin Zhang, Zihan Liu, Zhangyang Gao, Jiangbin Zheng, Stan.Z
AAAI, 2024
[PDF]
[BibTeX]
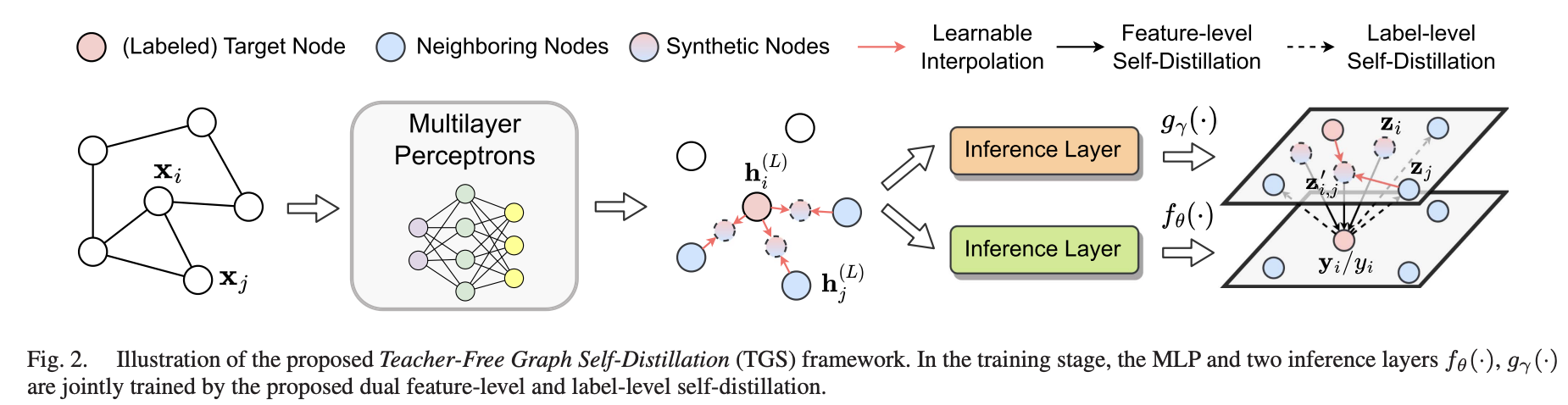
A Teacher-Free Graph Knowledge Distillation Framework With Dual Self-Distillation
Lirong Wu, Haitao Lin, Zhangyang Gao, Guojiang Zhao, Stan.Z
TNNLS, 2024
[PDF]
[Github]
[BibTeX]
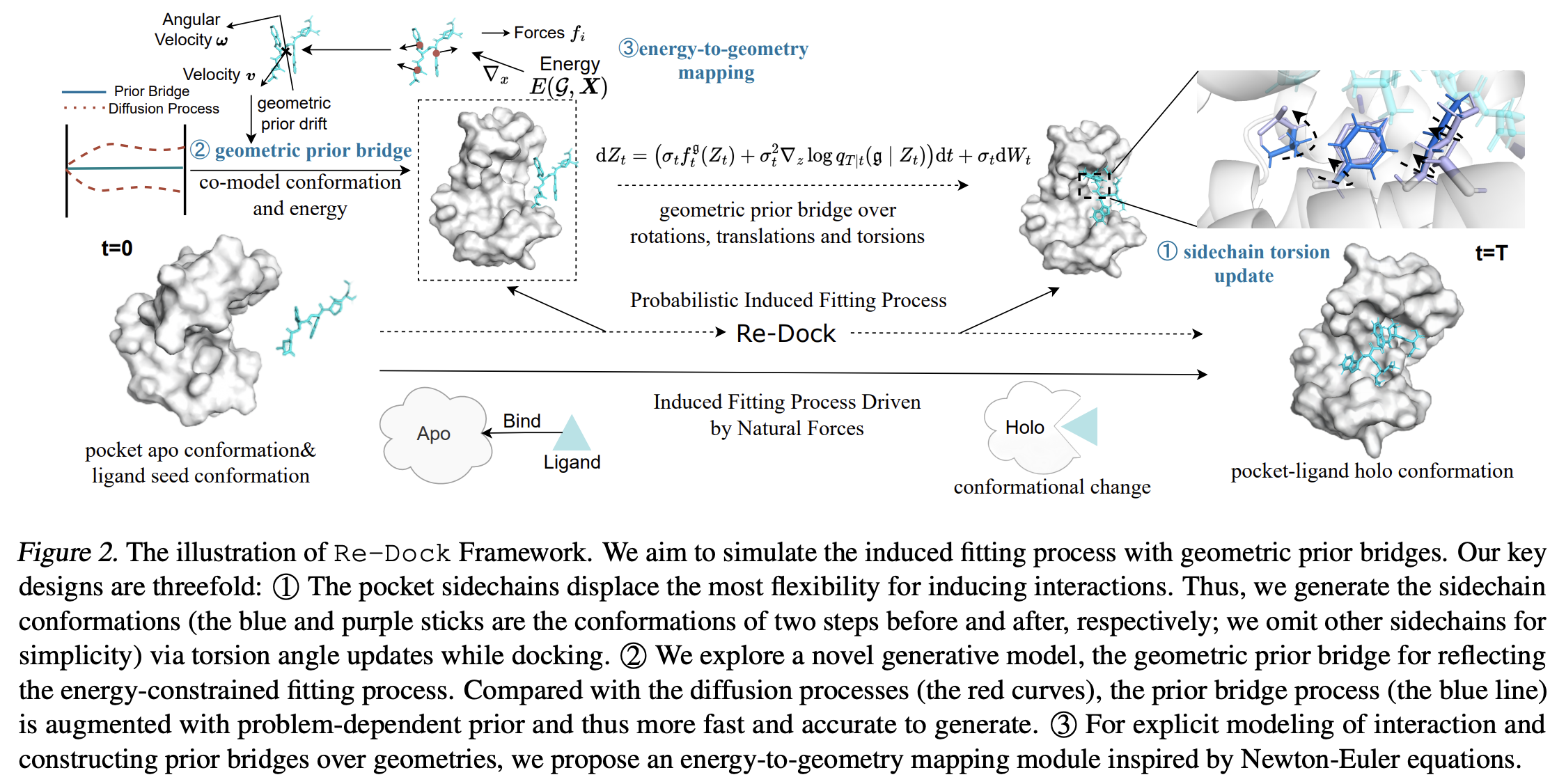
Re-Dock: Towards Flexible and Realistic Molecular Docking with Diffusion Bridge
Yufei Huang, Odin Zhang, Lirong Wu, Cheng Tan, Haitao Lin, Zhangyang Gao, Siyuan Li, Stan.Z
ICML, 2024 (Spotlight)
[PDF]
[BibTeX]
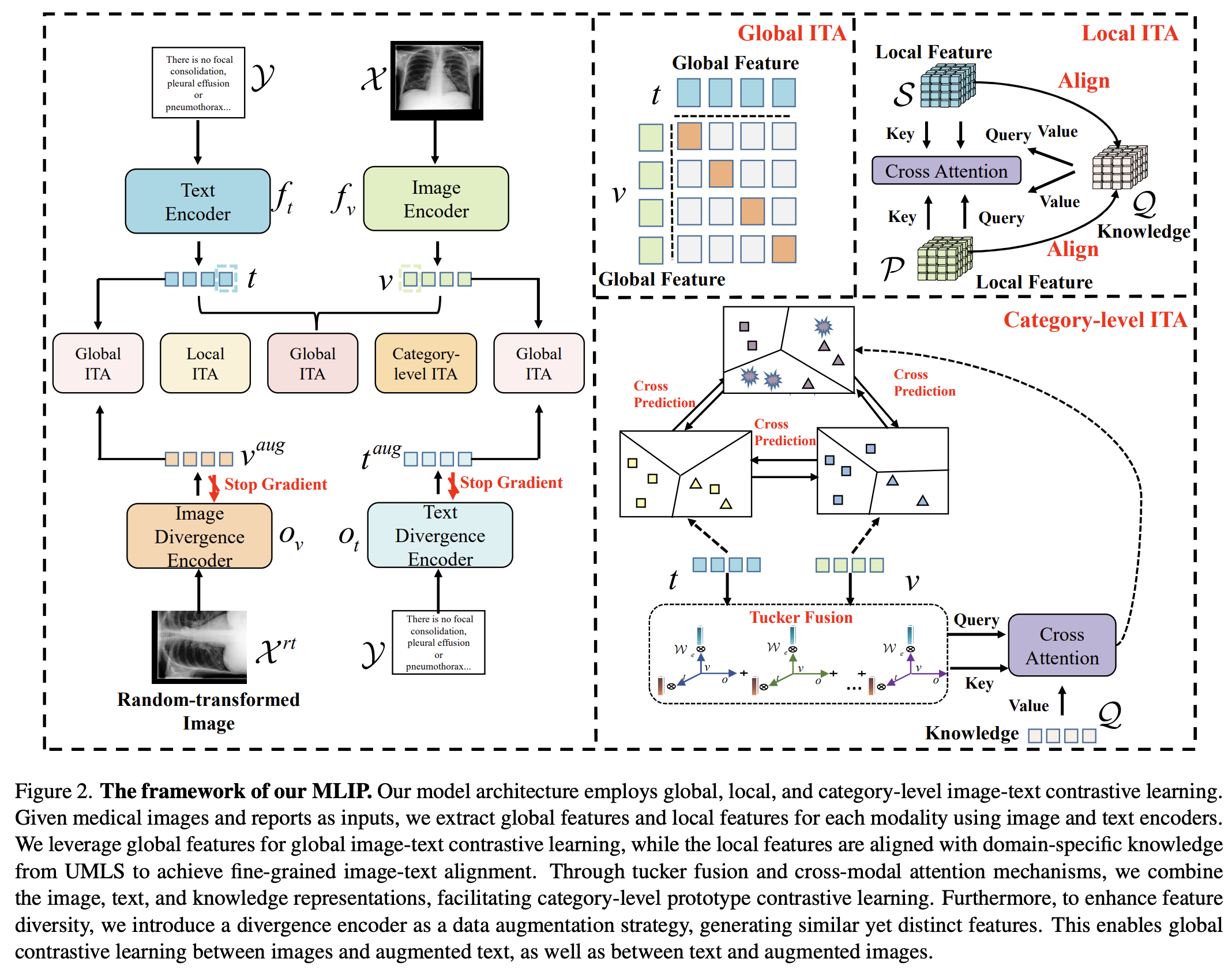
Mlip: Enhancing medical visual representation with divergence encoder and knowledge-guided contrastive learning
Zhe Li, Laurence T. Yang, Bocheng Ren, Xin Nie, Zhangyang Gao, Cheng Tan, Stan Z.
CVPR, 2024
[PDF]
[BibTeX]
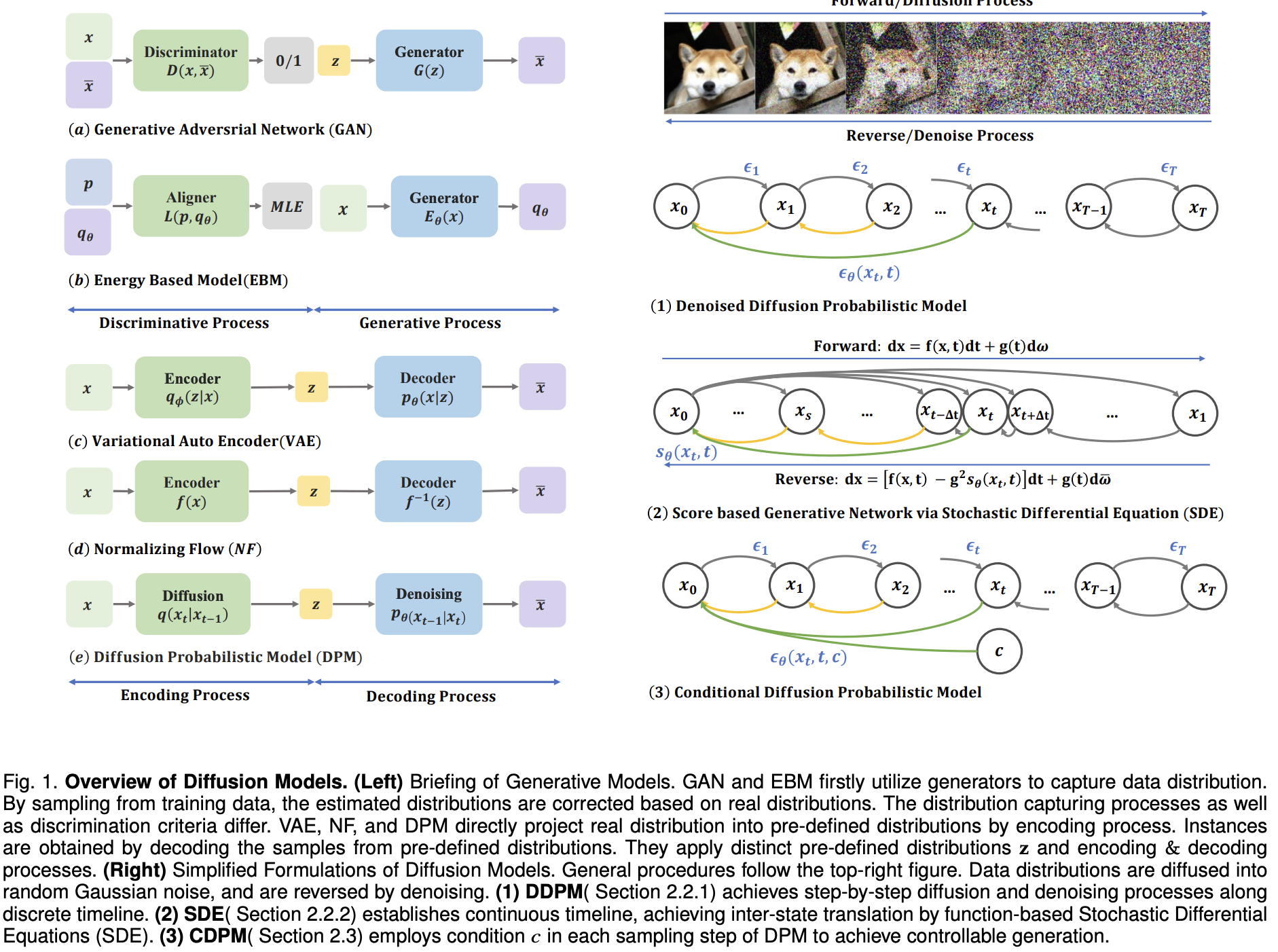
A Survey on Generative Diffusion Models
Hanqun Cao, Cheng Tan, Zhangyang Gao, Yilun Xu, Guangyong Chen, Pheng-Ann Heng, Stan Z.
TKDE, 2024
[PDF]
[Github]
[BibTeX]
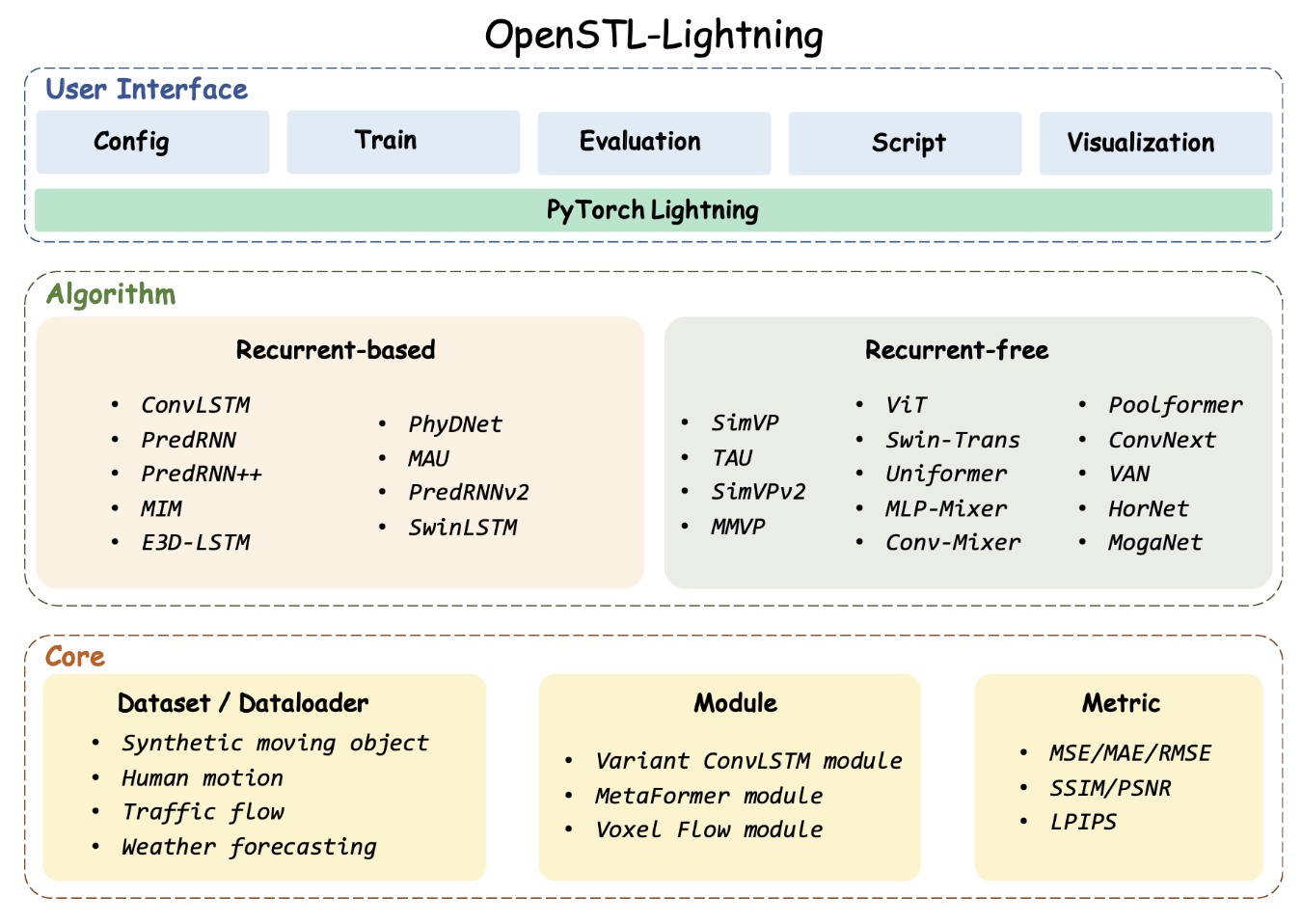
Openstl: A comprehensive benchmark of spatio-temporal predictive learning
Cheng Tan, Siyuan Li, Zhangyang Gao, Wenfei Guan, Zedong Wang, Zicheng Liu, Lirong Wu, Stan Z.Li
NeurIPS, 2023
[PDF]
[Github]
[BibTeX]
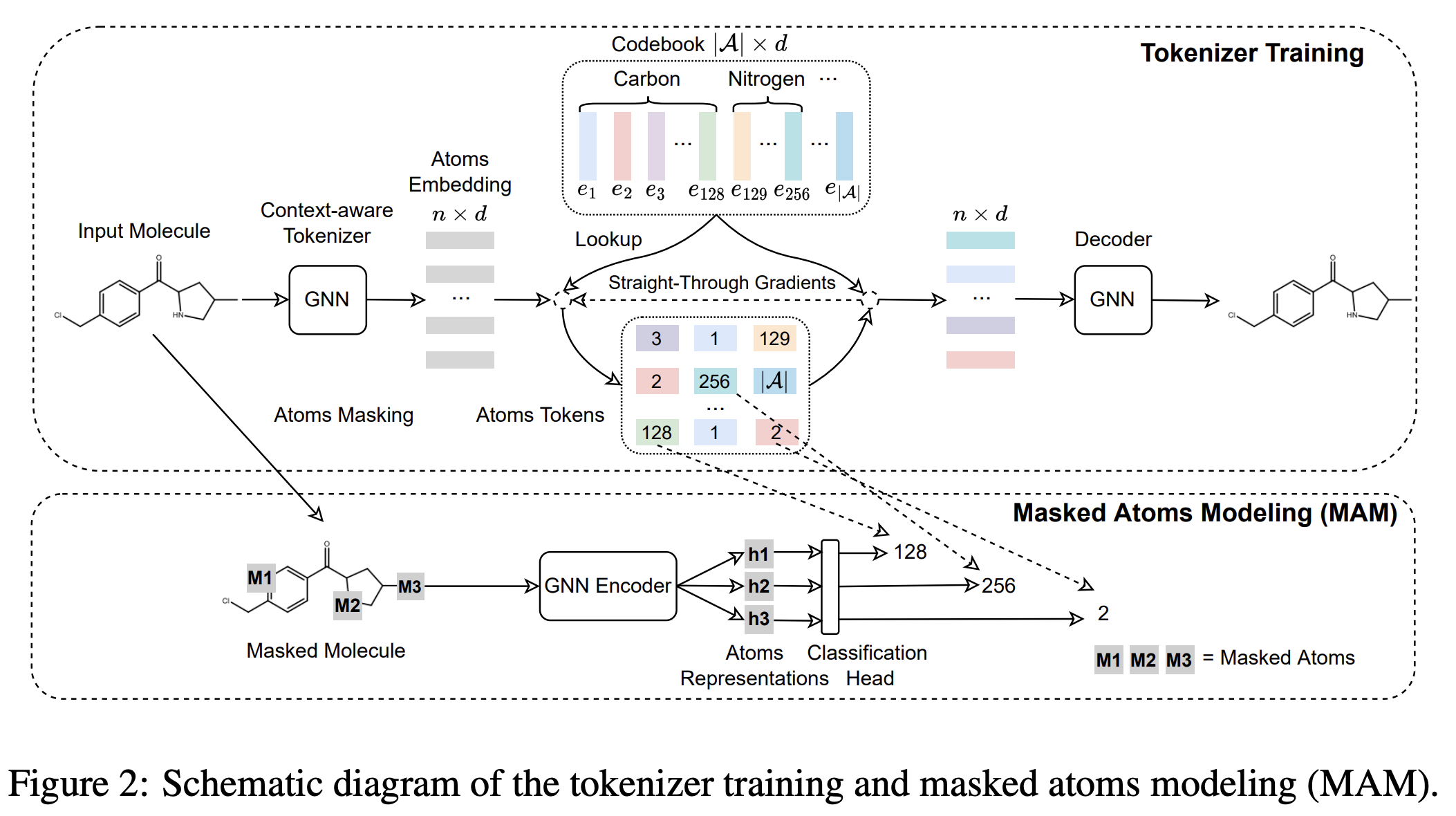
Mole-bert: Rethinking pre-training graph neural networks for molecules
Jun Xia, Chengshuai Zhao, Bozhen Hu, Zhangyang Gao, Cheng Tan, Yue Liu, Siyuan Li, Stan Z.Li
ICLR, 2023
[PDF]
[Github]
[BibTeX]
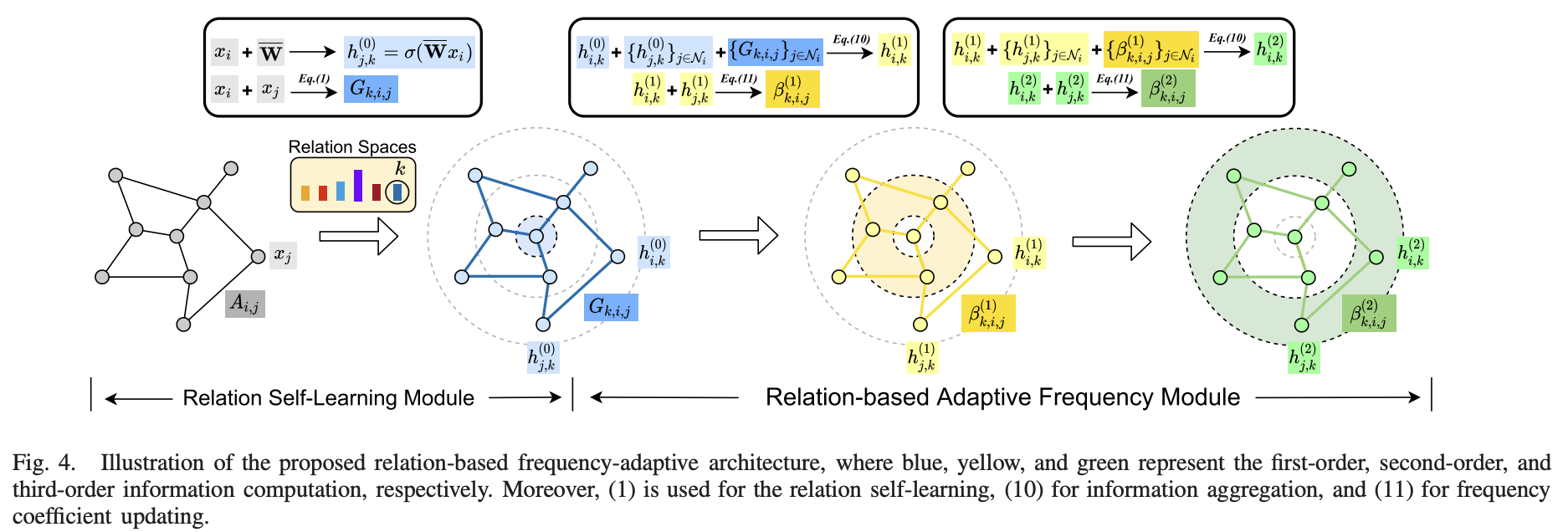
Beyond homophily and homogeneity assumption: Relation-based frequency adaptive graph neural networks
Lirong Wu, Haitao Lin, Bozhen Hu, Cheng Tan, Zhangyang Gao, Zicheng Liu, Stan Z.Li
TNNLS, 2023
[PDF]
[Github]
[BibTeX]

Self-supervised learning on graphs: Contrastive, generative, or predictive
Lirong Wu, Haitao Lin, Cheng Tan, Zhangyang Gao, Stan Z.Li
TKDE, 2021
[PDF]
[Github]
[BibTeX]
Services
Program committee member | Reviewer
- IEEE Conference on Computer Vision and Pattern Recognition (CVPR)
- International Conference on Computer Vision (ICCV)
- International Conference on Learning Representations (ICLR)
- Conference and Workshop on Neural Information Processing Systems (NeurIPS)
- International Conference on Machine Learning (ICML)
- Association for the Advancement of Artificial Intelligence (AAAI)